Soon after contract discussions have started with the WCIC Associate Director, clients next work with the Molecular Technologies Department (MTD) for all plant transformation projects conducted at the WCIC. The MTD provides comprehensive project and plasmid design, with Golden Gate mediated assembly of plant transformation DNA molecules as an output, which is unique amongst public plant transformation service providers. MTD produces transcriptional units which are comprised of synthetic DNA molecules which are compatible with the MoClo framework:
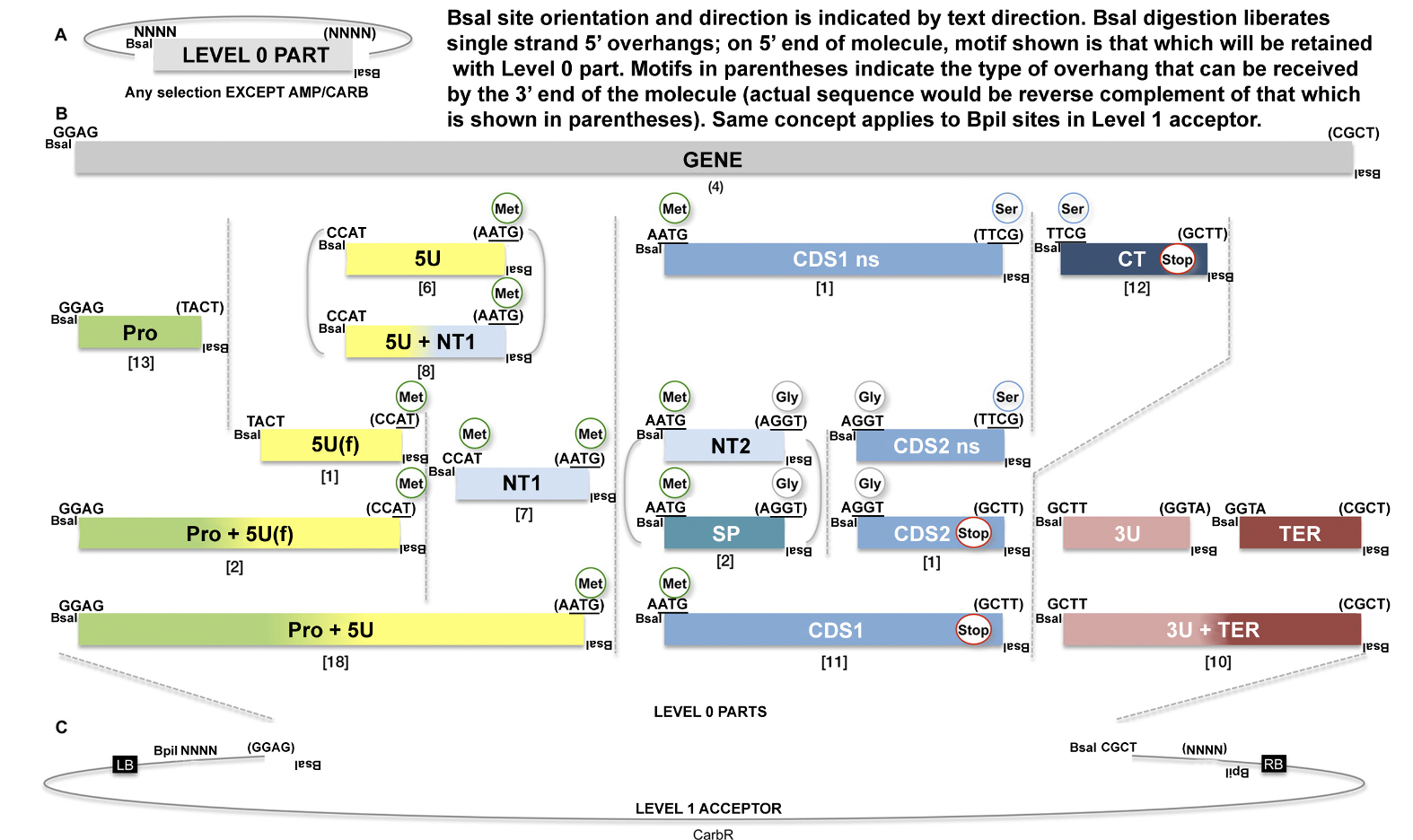
In addition to the basic overexpression and promoter reporter approaches, which are possible using the MoClo and MoClo Plant Parts Kits (Addgene Kits #1000000044 & #1000000047), we have designed the HARBOR System of Golden Gate tools, which make use of a number of the chromogenic proteins previously reported by the iGEM Uppsala Team (https://2013.igem.org/Team:Uppsala/chromoproteins) (See Figure 2 for WCIC-MTD examples of these useful tools).
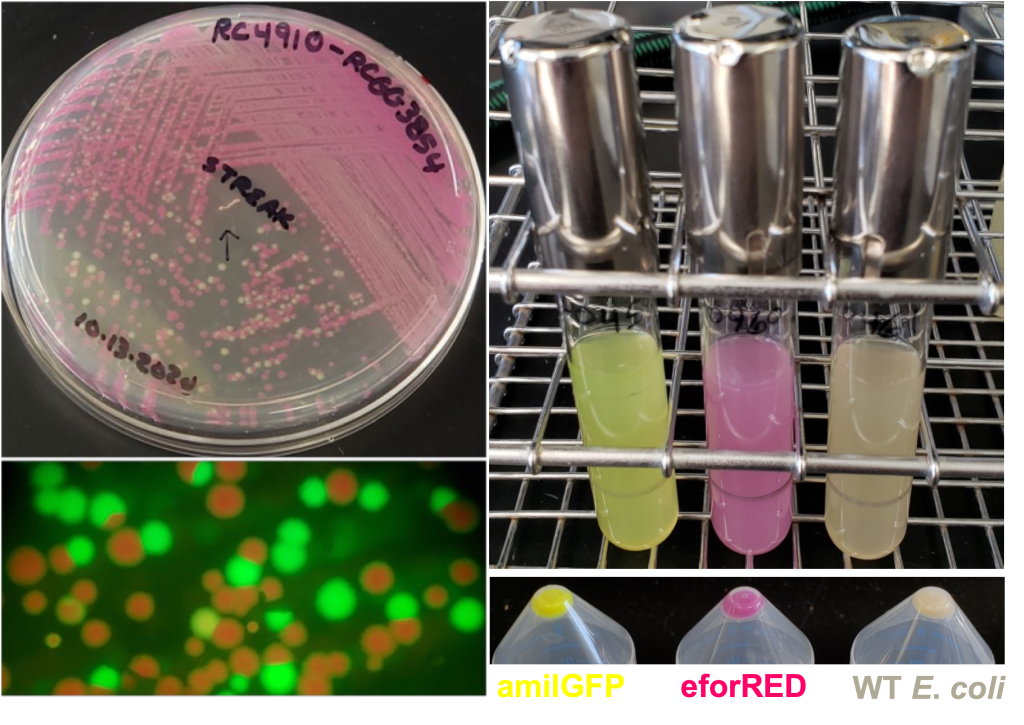
Figure 2. Examples of the iGEM Uppsala Team Chromogenic proteins used in the HARBOR System plasmids. Acquisition or loss of these bacterially expressed scorable markers is a key component to making the WCIC Plasmid Assembly system efficient.
Each of the subsystems of the HARBOR system have been designed to generate, as outputs, DNA molecules which dovetail seamlessly into the aforementioned MoClo framework (see Figure 1). The HARBOR subsystems help to expand what is scientifically possible with the MoClo kits and enable the following advanced plant molecular biology approaches:
| HARBOR SUBSYSTEM | SCIENTIFIC APPROACH |
|---|---|
| BLACKSMITHv4 | Polycistronic gRNA (tRNA delineated) for CRISPR/Cas9 (see Xie, Minkenberg, and Yang, PNAS (2015) v112 n11 p3570-3575. https://doi.org/10.1073/pnas.1420294112 |
| BLACKSMITHv6 | Polycistronic gRNA (tRNA delineated) for Phloem mobile CRISPR/Cas9 (see Yang et al., Nature Biotechnology (2023) v41 p958-967. https://doi.org/10.1038/s41587-022-01585-8 |
| DAGGERv2 | amiRNA (see Carbonell et al., The Plant Journal (2015) v82 n6 p1061-1075. https://doi.org/10.1111/tpj.12835 |
| HARPOONv3 & v4 | RNAi (v3 for monocots, v4 for dicots) |
| RIGGERv4 | Polycistronic messages (see Figure 3) |
| EXPLODER-II | Backbone free, linear DNA for bombardment |
| Two T-DNA | DICOT and MONOCOT strategies in binary plasmid and GAANTRY system formats |
| SeedRUBYv1 | Soybean, Maize, Sorghum, Barley, Wheat, Brachypodium (most frequently used in the EDIT constructs) (see Figures 4 & 5) |
| DIRECTCLONETM | DICOT and MONOCOT CRISPR/Cas9, Promoter GUS, Constitutive OE, Terminator tests, Intron Mediated Enhancement (IME) – One Golden Gate mediated cloning step to completed plant transformation plasmid (see Figures 6 & 7). |
| GAANTRY | In addition to enabling the production of very large T-DNA molecules (we currently impose an upper limit of 85000 basepairs) for dicots, we exclusively use GAANTRY for all DICOTEDITTM constructs (binary plasmids no longer built for dicot editing) (see Figure 8). Additionally, two T-DNA strategies are available for the GAANTRY approach. We are actively working to bring GAANTRY technology online for our monocot pipelines. We use Golden Gate mediated assembly to build all of our GAANTRY compatible CONTAINERS, which means that our entire synthetic parts collection can be used for this approach. For a nominal fee the entire GAANTRY strain genome can be sequenced to provide assurance that the ~200 Kbp virulence plasmid sequence (containing your T-DNA) is exactly as designed. |
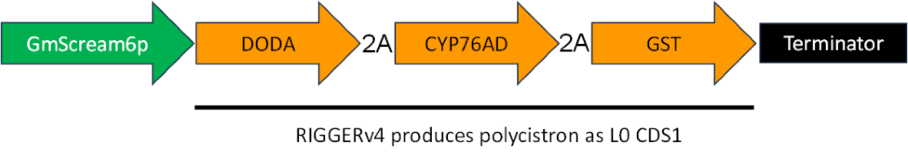
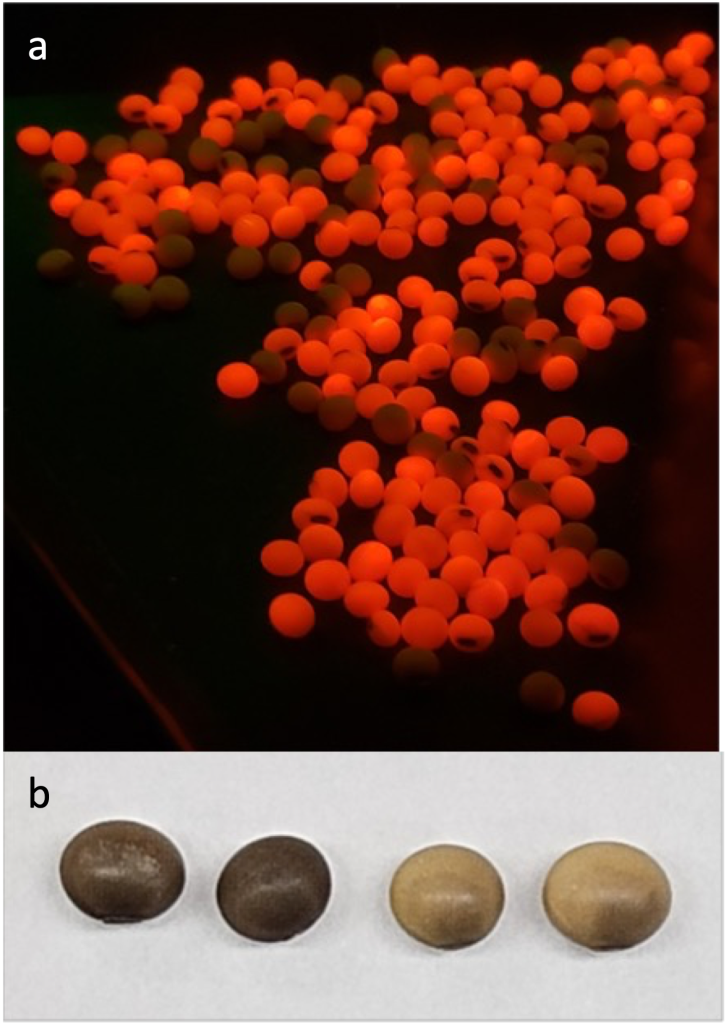

Figure 5. (Top) SeedRUBYv1 in MONOCOTS. The RUBYv1 polycistron (assembled in house using the RIGGERv3 System) under control of either the Ta1Dy10 promoter (Thilmony et al., (2014) GM Crops Food v5 n1 p36-43) for either Brachypodium distachyon or in wheat, the D-hordein promoter (Cho et al., (2002) Physiologia Plantarum v115 p144-154) for barley, the a–kafirin promoter (Liu et al., Plant Cell Tiss Organ Cult (2017) v128 p133-143) for sorghum, and the a–zein promoter (Requesens et al., (2019) Transgenic Res v28 p537-547) for maize.

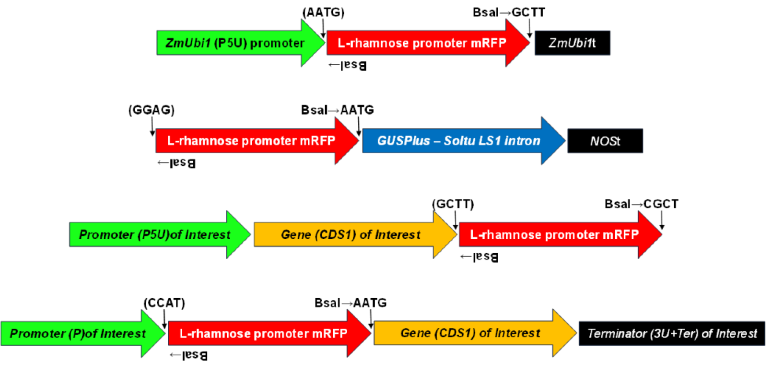
Figure 8. The GAANTRY System (Collier, Thomson, and Thilmony et al., (2018) Plant J v95 n4 p573-583) for in vivo, in situ assembly of T-DNA molecules via recombinase mediated cassette exchange (RMCE). All gene editing projects at the WCIC are now conducted exclusively in ArPORT1 clones generated using the GAANTRY system (virulence plasmid T-DNA launch, no binary plasmid). THERE IS NO BINARY PLASMID!
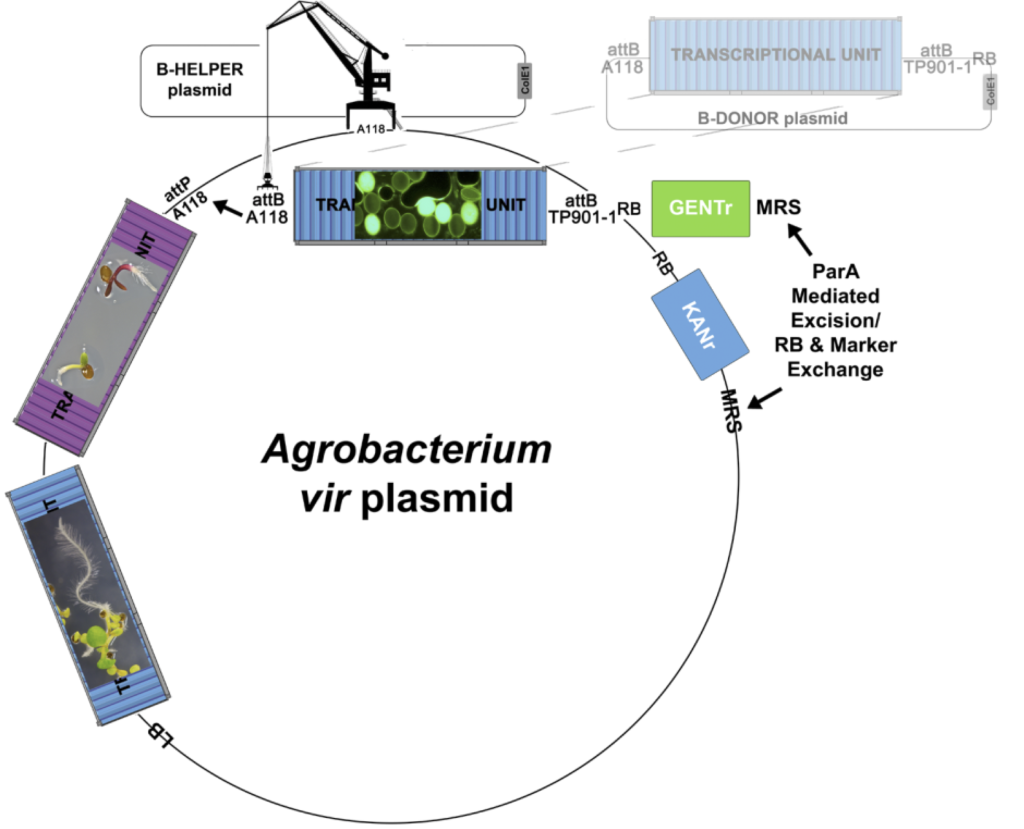
We build plasmids and GAANTRY strains exclusively for plant transformation projects conducted at the WCIC, we do not build materials for external use.
For those clients who choose not to take full advantage of our plasmid design and assembly service, underpinned by the large and growing WCIC Golden Gate Synthetic Parts repository, it may be possible to use a plant transformation plasmid which you have designed and assembled in your lab. To ensure the best chance for project success, we recommend that you consult with us prior to construct assembly to ensure that the choices made regarding plant selection marker are compatible with our standard plant transformation protocols. For those clients whose materials can be accepted, we require the plasmid to be ethanol precipitated and dried (please do not use a Speedvac or lyophilizer):
DNA submission instructions:
For each construct make a full mini preparation of plasmid DNA (typically 5 to 10 mL of liquid culture). Ethanol precipitate (add 1/10th volume 3M NaOAc + 2 volumes of 100% EtOH) then pellet the DNA by centrifugation. Remove the EtOH and fully dry the pellet. When dry, seal the clearly labeled (we also prefer that the labeling is protected with an overlay of Scotch brand (3M) Transparent tape) tube with Parafilm. Ship the sealed tube in a hardened container (either a hardsided box, or place the tube inside a 50 mL polypropylene tube, which then goes in a box) to:
Molecular Technologies Department
Wisconsin Crop Innovation Center
8520 University Green
Middleton, WI 53562
